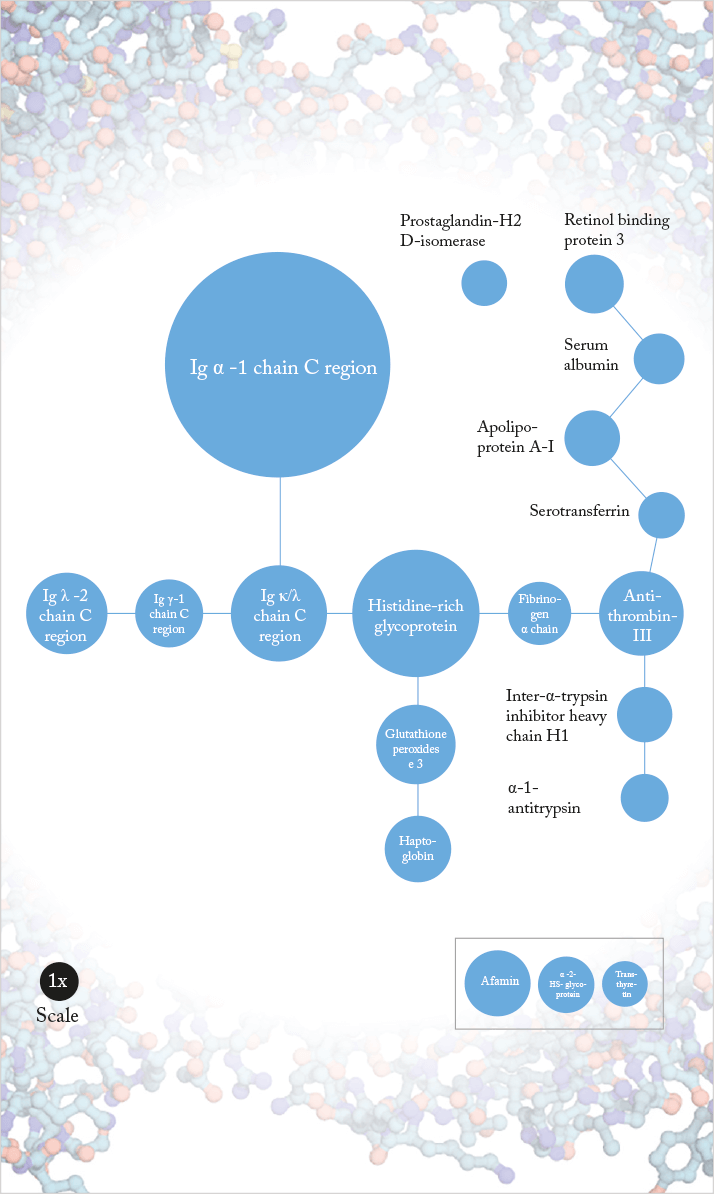
To gain a better understanding of the pathophysiology of wet AMD, we analyzed the proteomes of 88 vitreous samples (73 from patients with AMD and 15 patients with idiopathic floaters serving as controls) (1). After vitreous sampling, we used mass spectrometry to detect, sequence and identify the proteins present. Statistical analysis revealed 19 proteins (Figure 1) with significantly increased abundance in AMD. Most of these proteins are secreted, with functions that include biological transport, fatty acid binding, protease inhibition and processes involving hydrogen peroxide. They form part of a densely interconnected network that includes an immunoglobulin cluster (suggesting a role in inflammatory responses), and processes including cell adhesion, lipid metabolism, apoptosis prevention and regulation of proteolysis.
Michael Koss is currently the head of the retina unit of the Department of Ophthalmology at the oldest German university in Heidelberg.
References
- MJ Koss et al., “Proteomics of vitreous humor of patients with exudative age-related macular degeneration”, PLoS One. 9, e96895 (2014). PMID:24828575.
