Autophagy – an intracellular degradation system that delivers cytoplasmic contents to the lysosome for breakdown and recycling – is one of those simple processes in cell biology that actually plays a wide variety of physiological and pathophysiological roles (1). And it turns out that mutations in an autophagy-related gene expressed in the retinal pigment epithelium (RPE) are the cause of a newly identified type of adult-onset retinal dystrophy with early macular involvement.
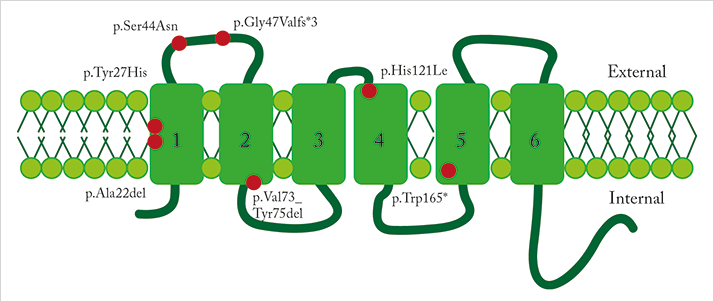
Results published in the American Journal of Human Genetics (2) describe people from five families with a variety of DRAM2 mutations (Figure 1), all of which led to the loss of central vision from the ages of ~30–40 years onwards, and also the loss of peripheral vision in older individuals. DRAM2, or to give it its full name, DNA damage-regulated autophagy modulator protein 2, encodes a transmembrane lysosomal protein that plays a role in initiating autophagy, and is expressed in both photoreceptors and the surface of RPE cells. One of the paper’s co-authors and leader of the team that made the initial discovery, Manir Ali, noted that, “a high level of autophagy takes place in RPE due to the need for constant renewal of the photoreceptor outer segments following daily light-induced damage”, explaining that “it is therefore likely that, in the absence of correctly functioning DRAM2 protein, autophagy and photoreceptor renewal is reduced, leading to thinning of the photoreceptor cell layer. Our findings suggest that DRAM2 is essential for photoreceptor survival.”
References
- N Mizushima, “Autophagy: Process and Function”, Genes & Dev, 21, 2861–2873 (2000). PMID: 18006683. ME El-Asrag, et al., “Biallelic mutations in the autophagy regulator DRAM2 cause retinal dystrophy with early macular involvement”, Am J Hum Genet, 96, 695–708 (2015). PMID: 25983245.
